Biography
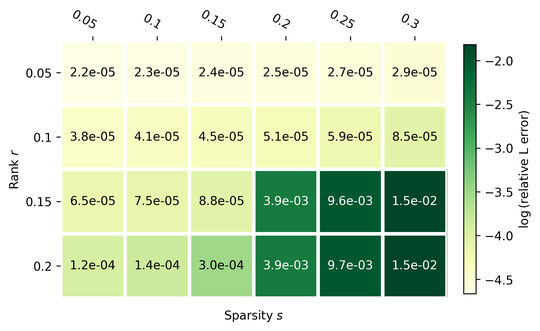
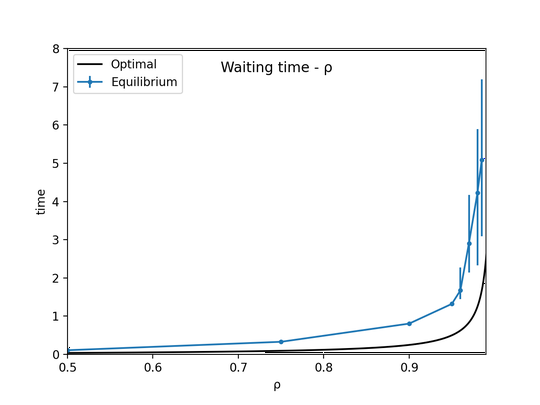
I am currently a third year PhD student at CMS department, Caltech, where I am very fortunate to be advised by Yisong Yue and Yang Song. My research focuses on generative modeling, in particular inference-time posterior sampling algorithms with pretrained diffusion priors, with application to reward guided generation and solving scientific inverse problems. More recently, I have been working on developing more generation-friendly visual tokenizers for latent generative models.
I like playing piano and I especially enjoy playing the songs from animes. I am also a fan of chess and photography.
Download my curriculum vitae.
- Generative Models
- Inverse Problems
-
California Institute of Technology, 2023-Present
Computing + Mathematical Science
-
Tsinghua University, 2019-2023
Yao Class, Institute for Interdisciplinary Information Sciences
-
Shenzhen Middle School, 2013-2019
| Sep, 2025 | Two papers accepted to NeurIPS 2025! Meet me at San Diego :) | |
|---|---|---|
| April, 2025 | Our paper DAPS received an oral presentation at CVPR 2025! | |
| Jan, 2025 | Our paper InverseBench received a spotlight presentation at ICLR 2025! | |
| Sep, 2023 | I joined Caltech CMS as a PhD student! | |
| June, 2023 | I graduated from Tsinghua University! |
Publications
Experience
I was fortunate to be advised by Prof. Bo Li. Projects include:
- Certified robustness for point cloud models
- Certified robustness for multi-sensor fusion systems
- Fair federated learning by EM clustering
- I was fortunate to be advised by Prof. Xiaolin Hu and researched on launching physically realizable adversarial attacks on object detection models.